 |
|||||
|
Typing tool
|
Both noroviruses and sapoviruses are genetically and antigenically highly diverse.
The non-enveloped virus particles are 27-40 nm in diameter and have a single-stranded
positive-sense RNA genome of 7.1-7.7 kb in length. In human noroviruses, this genome
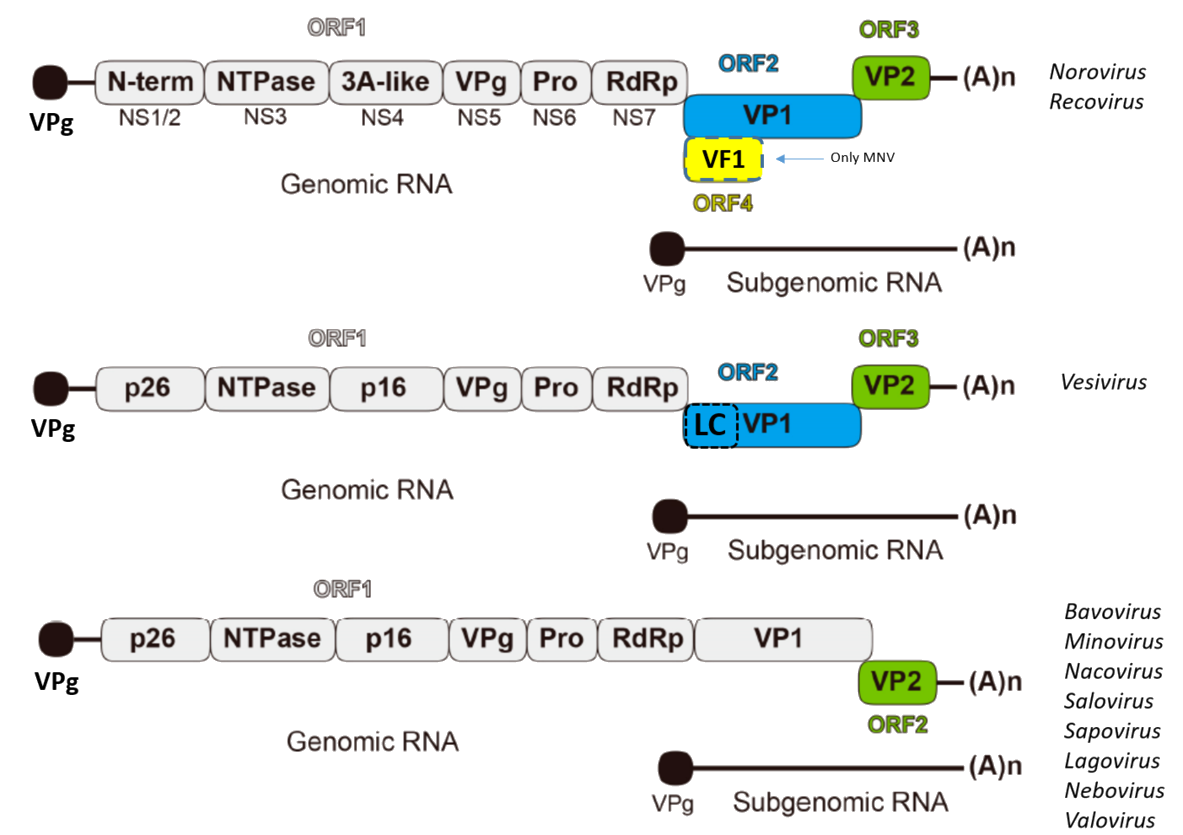
is divided into three open reading frames (ORF1-3)
whereas sapoviruses consist of 2 ORFs (see Figure).
Based on phylogenetic clustering of complete VP1 amino acid sequences, noroviruses can
now be classified into 10 established (GI - GX) and 2 tentative new norovirus genogroups,
of which viruses from GI, GII, GIV, GVIII and GIX (formerly GII.15) infect
humans [2].
Each genogroup can be further divided into genotypes (9 GI, 26 GII, 2 GIV, 1 GVIII, and 1 GIX).
In addition, for GI and GII at least 14 GI polymerase (GI.P) types and 37 GII.P types have been
described based on clustering of nucleotide sequences of the polymerase region [2].
Most laboratories amplify relative short sequences spanning the 3’-end of ORF1 (polymerase region)
and 5’-end of ORF2 (major capsid protein gene) to genotype GI and GII noroviruses.
These sequences can be generated in separate RT-PCR reactions or in a single,
genogroup-specific, RT-PCR assay [3].
This human calicivirus typing tool [4] uses a BLAST-like algorithm to type both the polymerase
region (P-type) and the capsid (genotype) of GI, GII, GIV, GVIII, and GIX (formerly GII.15)
noroviruses as well as capsid genotyping of GI, GII, GIV and GV sapoviruses [5]
against a database of norovirus and sapovirus reference sequences.
Genotypes are assigned following classification criteria agreed upon by the Norovirus
Classification Working Group [2].
Norovirus and Sapovirus
The family Caliciviridae consist of 11 genera [1] of which viruses belonging
to the Norovirus and Sapovirus genera cause epidemic and endemic
acute gastroenteritis (vomiting and diarrhea) in people of all ages.
The clinical symptoms of norovirus and sapovirus gastroenteritis are indistinguishable.
Reference sequences
The reference sequences used in this typing tool are updated on a regular basis.
They are listed under
Reference sequences tab
and are all available in GenBank.
Citations:
pdf
1.
Vinjé J, Estes MK, Esteves P, Green KY, Katayama K, Knowles NJ,
L'Homme Y, Martella V, Vennema H, White PA, ICTV Report Consortium.
ICTV Virus Taxonomy Profile: Caliciviridae.
J Gen Virol. 100(11):1469-1470 (2019).
2.
Chhabra P, de Graaf M, Parra G, Chan MC-W, Green KY, Martella V, Wang Q, White PA, Katayama K,
Vennema H, Koopmans MPG, Vinjé J.
Updated classification of norovirus genogroups and genotypes.
J Gen. Virol. 100 (10):1393-1406 (2019).
pdf
3.
Chhabra P, Browne H, Huynh T, Diez-Valcarce M, Barclay L,
Kosek MN, Ahmed T, Lopez MR, Pan CY, Vinjé J.
Single-step RT-PCR assay for dual genotyping of GI and GII norovirus strains.
J Clin Virol. 134:104689 (2021).
pdf
4.
Tatusov RL, Chhabra P, Diez-Valcarce M, Barclay L, Cannon JL, Vinjé J,
Human Calicivirus Typing tool: A web-based tool for genotyping human norovirus and sapovirus sequences.
J Clin Virol. 134:104718 (2021).
pdf
5.
Oka T, Wang Q, Katayama K, Saif LJ,
Comprehensive review of human sapoviruses.
Clin Microbiol Rev. 28 32-53 (2015).